Research Projects
The Prediction of NMR spin relaxation by Molecular dynamics
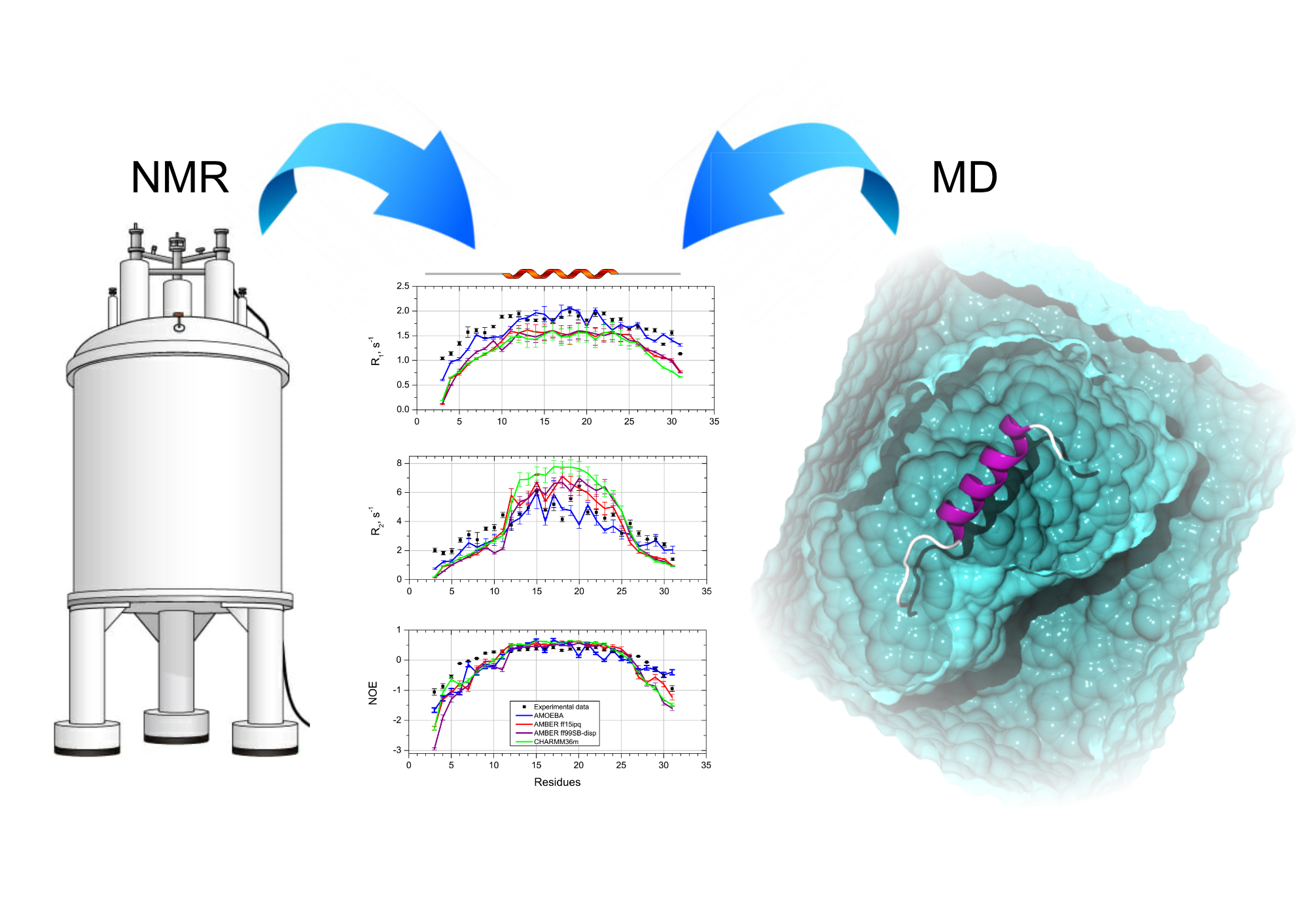
Dynamics is essential for proteins to perform interactions with biological partners. Among the vast landscape of biophysical methods, NMR possesses this exquisite potential to sample motions at different frequency through the measurement of various relaxation parameters. The interpretation of relaxation data is commonly subjected to the choice of a suitable model, like the model free approach, for instance. We are seeking to develop an approach based on molecular dynamics to predict all the available NMR spin relaxation. See references related to the use of quaternion to predict the rotational diffusion tensor, the prediction of R1, R2 and NOEs and the use of polarizable force fields.
Elucidating the bacterial silver resistance
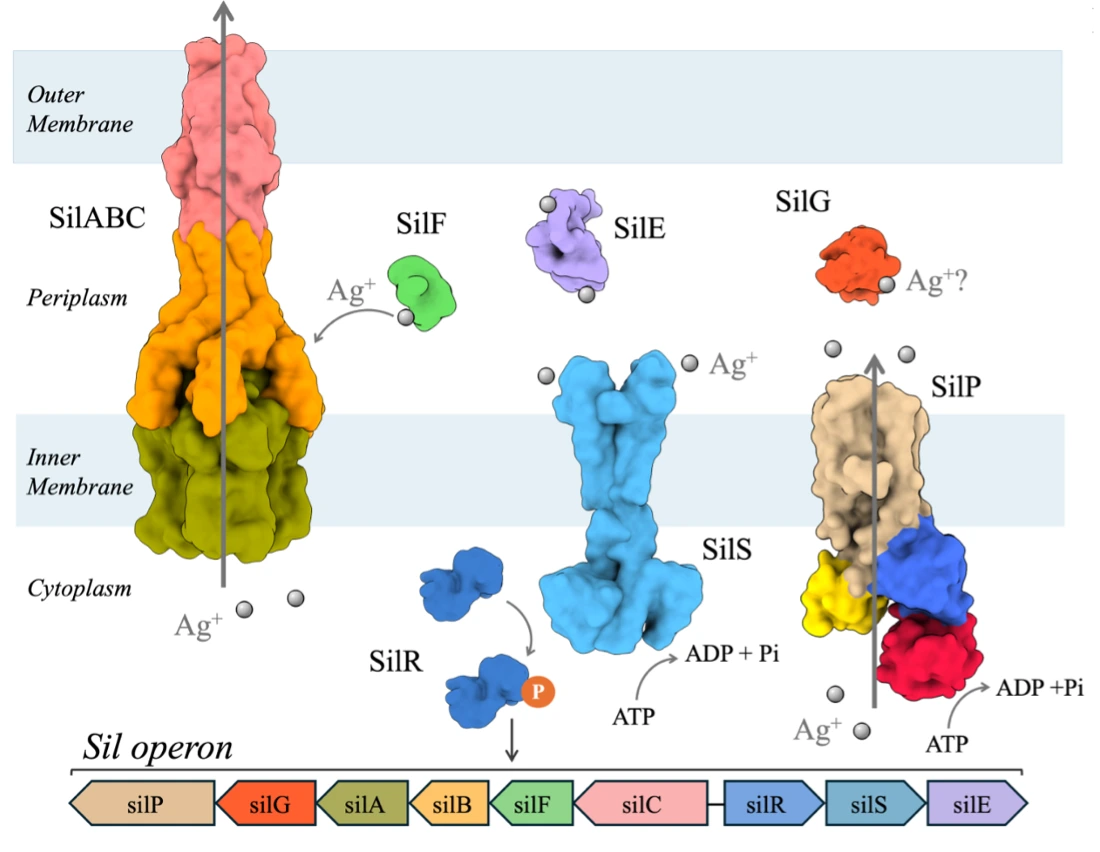
Silver antimicrobial properties have been used for thousands of years, from the Chaldeans as early as 4000 B.C.E. Despite this long-standing history and its demonstrated activity against Gram-negative bacteria, the complete bactericidal mode of action of silver remains unclear. To counteract the toxic effect of silver, Gram-negative bacteria have developed different resistance mechanisms, including the efficient transport of the metal out of the cell promoted by an efflux pump. The latter one is encoded by the silver-resistant gene cluster named sil composed of 9 proteins: an Ag+/H+ tripartite efflux pump (SilABC), a P-type ATPase efflux pump (SilP), a two-component regulatory system (SilRS) and three periplasmic silver-binding proteins SilE, SilG and SilF. First, our group investigated the structures of SilE mimicking peptides upon silver binding, then we provided structural and dynamical insights into full-length Ag+-bound SilE from combined analytical probes. Our group also made a step forward with the structural and dynamical characterization of the SilF/SilB complex, where they unraveled the mechanism by which SilF transfers silver ions and interacts with SilB that belongs to the efflux pump SilABC.
Host-parasite interactions – understanding immune evasion of parasitic worms
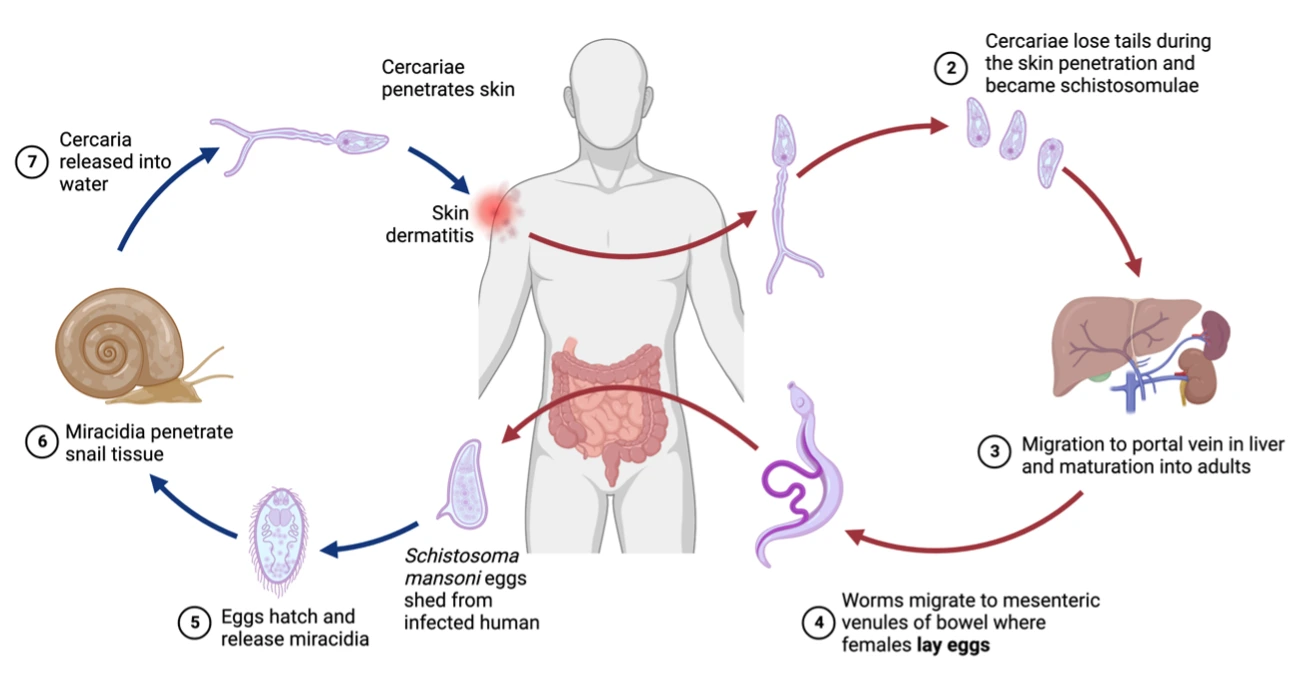
Parasitic trematodes of the genus Schistosoma are digenean with a complex life cycle, involving 2 free living stages and 2 hosts, a mollusk as intermediate and a mammal definitive one. Each life stage is adapted to its environment and modulates it in turn. We are interested in the macromolecules excreted and secreted by the stages infecting humans to understand their role in immune system modulation. In particular we focus on two families of highly variable proteins: Micro-exons genes (MEGs) and Venom antigen-like proteins (VALs). Both families have no enzymatic activities, are differently expressed during the lifetime and are particularly rich in Cys. On top of being transcribed by several genes, alternative splicing increases the variability and the number of isoforms for each gene product. We are producing in heterologous systems, candidates of both families and characterising their structure by means of macromolecular crystallography (MX), SAXS, NMR and also their interactions with circulating host partners and receptors by means of molecular-scale biophysics (DLS, SPR, fluorescence spectroscopy). Moreover, given their abundance in blood circulation, VALs have been validated as biomarkers and are going to be exploited in a point-of-care (POC) diagnostic test, following the WHO NTD-Roadmap 2030.
Functional dynamics of biomolecular motors
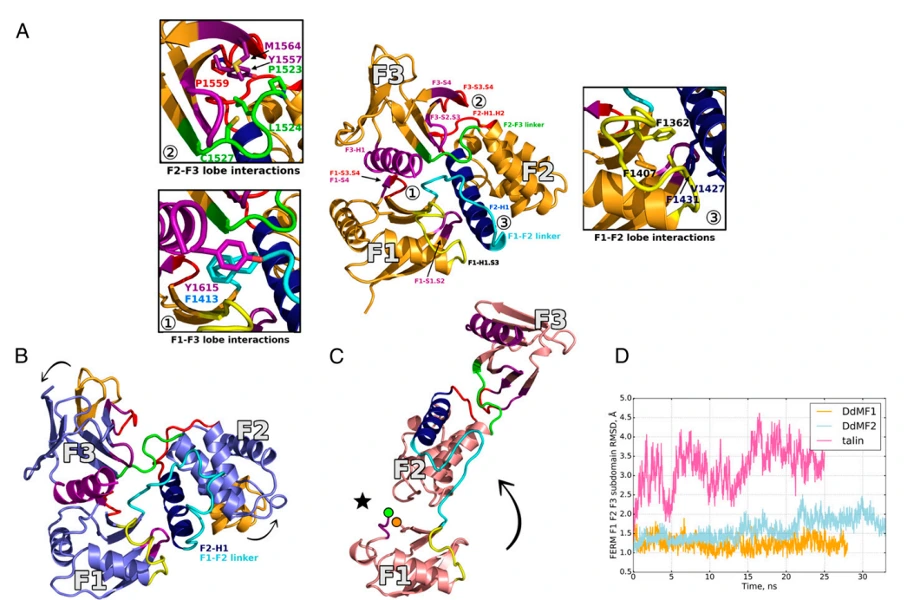
Biomolecular motors are proteins that convert the free energy of ATP hydrolysis into mechanical work. Myosins are an important family of molecular motors necessary for many critical tasks such as cargo transport, cell division, cell motility and muscular contraction. Defective or mutated myosins are responsible for serious diseases like hypertrophic cardiomyopathy and cancers. This project uses enhanced sampling MD simulations of myosins to illuminate the conformational transitions that underpin the motor mechanism, allowing us to understand the structural basis for free energy conversion. This also helps us guide in silico discovery of drugs active against myosins.
Scaffold-derived peptides to treat an aggressive infant cancer called neuroblastoma
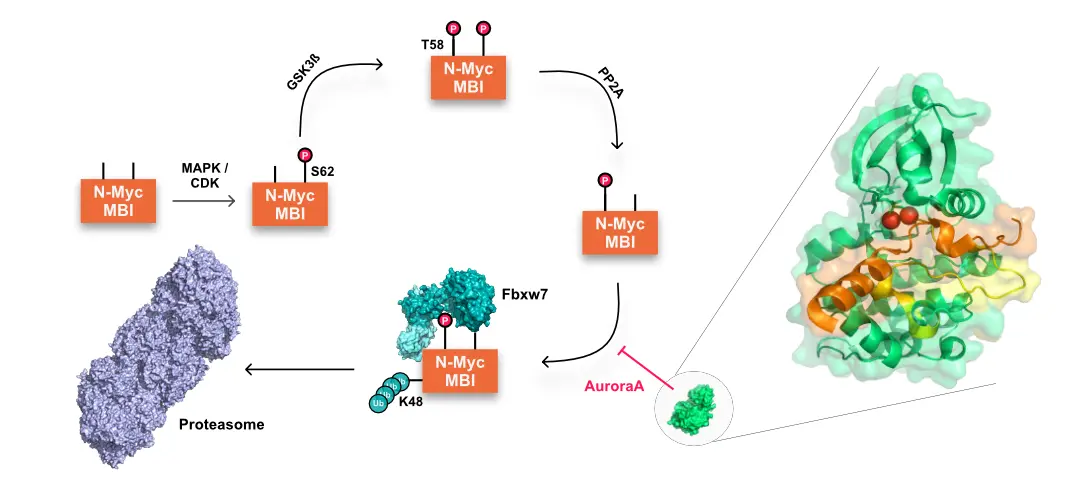
Neuroblastoma is a paediatric cancer characterised by a high mortality rate, where amplification of the MYCN oncogene determines the worst prognosis. This gene encodes N-myc, an intrinsically disordered protein (IDP) that acts as a transcription factor during neuronal development. The highest levels of N-myc are responsible for the cancerous transformation of neuronal cells. As it is difficult to eliminate N-Myc excess using classical drugs, we chose to target the AuroraA/N-myc complex, which once formed protects N-Myc from proteasomal degradation. Therefore, protein-protein interaction (PPI) inhibitors are designed in silico to disrupt this complex, allowing the proteasome machinery to eliminate N-Myc. These inhibitors are analysed using in vitro biophysical methods such as nuclear magnetic resonance (NMR), macromolecular crystallography (MX), small-angle X-ray scattering (SAXS), dynamic light scattering (DLS), circular dichroism and infrared spectroscopy. This project is in collaboration with Sapienza University of Rome where all the molecules designed are further tested in cellula.
 Biophysics of complex systems
Biophysics of complex systems